Mitochondria Length Classification AI-Powered
1. Pipeline: (3D) Mitochondria Length Classification AI-Powered
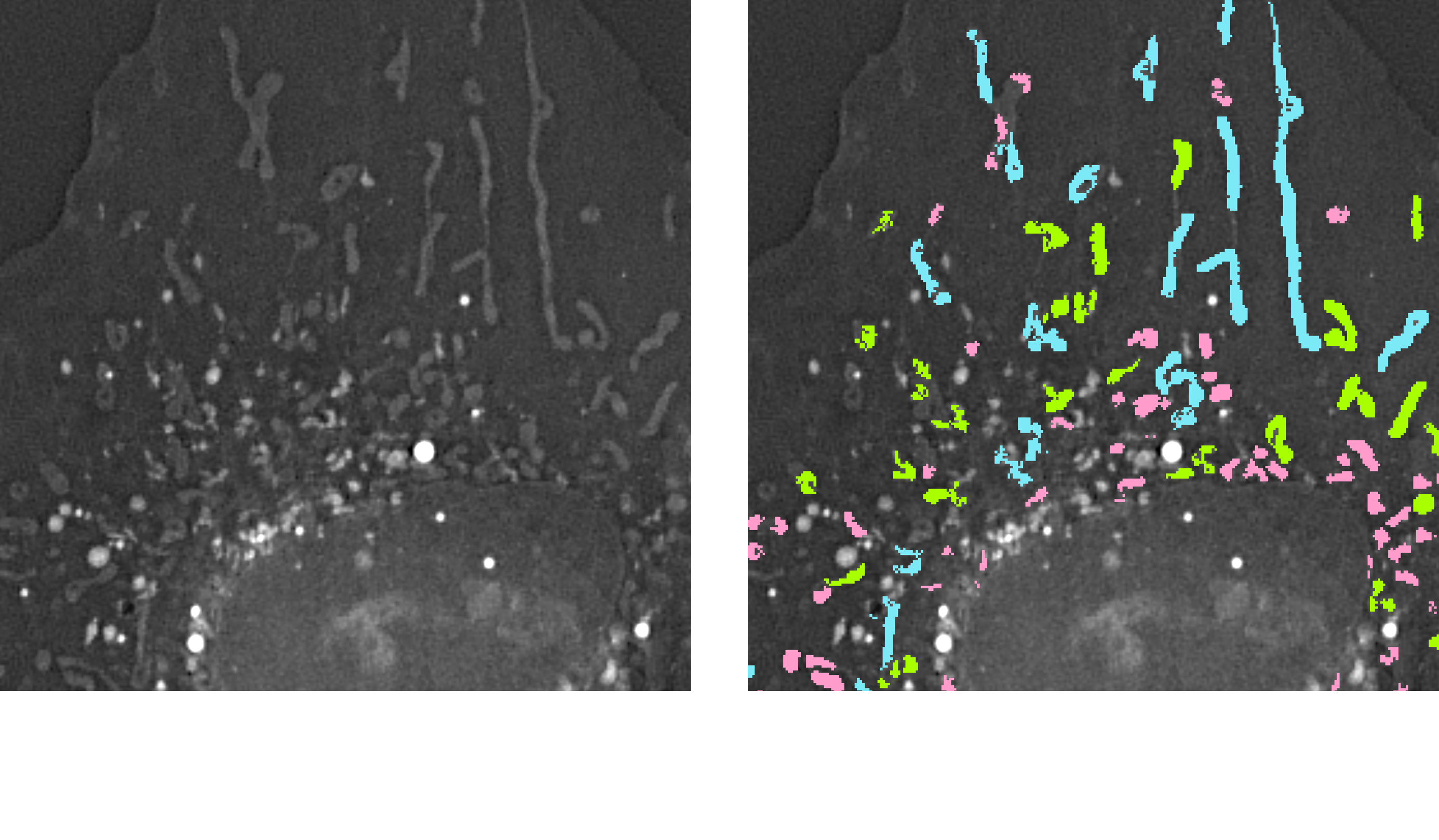
This pipeline classifies 3D mitochondria based on their morphological characteristics, specifically length, using AI-powered segmentation and rule-based filtering. Mitochondria are first segmented in 3D from holotomographic images, and then categorized into three morphological groups: elongated, intermediate, and fragmented, using the filter-by-measure process.
Each group is visualized as a distinct mask, allowing clear spatial interpretation of mitochondrial morphology within the cell population. The pipeline also provides quantitative analysis for each class, supporting studies on mitochondrial dynamics, fission and fusion balance, and cellular health status.
Output: 3D mitochondria masks classified by length (elongated, intermediate, fragmented), class-specific measurements (count, volume, mean length)
This pipeline performs per-cell lipid droplet (LD) analysis in 2D projections by combining rule-based LD segmentation with AI-powered cell segmentation. LDs are detected based on their high refractive index (RI) values and round morphology using a traditional rule-based approach. To enable cell-specific quantification, the Cellpose model is used to generate single-cell masks, allowing each droplet to be assigned to its corresponding cell. This integrated approach provides both object-level and per-cell LD measurements, making it ideal for studying cell-to-cell variation in lipid content. The pipeline is well-suited for high-throughput analysis, especially when cells are well-separated in 2D. Output: Single-cell masks, Labeled lipid droplets with individual measurements, Per-cell LD measurements
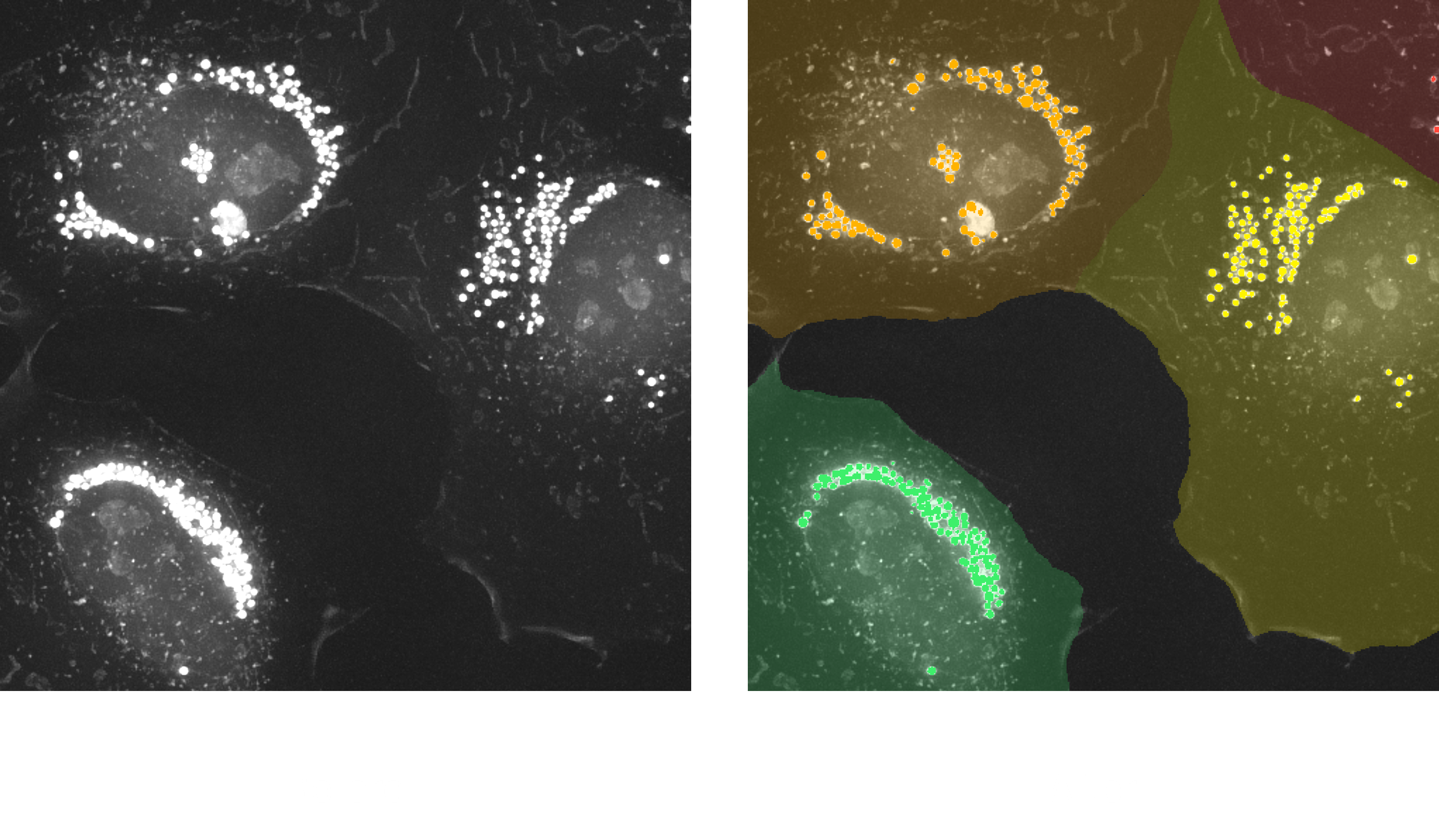
2. Pipeline: (2D) Lipid Droplet per Cell AI-powered
This pipeline performs per-cell lipid droplet (LD) analysis in 2D projections by combining rule-based LD segmentation with AI-powered cell segmentation. LDs are detected based on their high refractive index (RI) values and round morphology using a traditional rule-based approach. To enable cell-specific quantification, the Cellpose model is used to generate single-cell masks, allowing each droplet to be assigned to its corresponding cell.
This integrated approach provides both object-level and per-cell LD measurements, making it ideal for studying cell-to-cell variation in lipid content. The pipeline is well-suited for high-throughput analysis, especially when cells are well-separated in 2D.
Output: Single-cell masks, Labeled lipid droplets with individual measurements, Per-cell LD measurements
Access to analysis pipelines is available upon request. Please send your inquiry to info@tomocube.com to explore the options best suited to your needs.